iTools
iTools offers a graphical interface for perusing meta-data information about software tools, databases and material resources freely provided by NCBC funded institutions. iTools is built upon the XML framework prescribed by biositemaps.org, for generating and disseminating resource descriptions.
Visit Website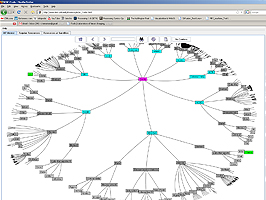

Features
- Produces, traverses and consumes Biositemaps
- Provides an interactive Hyperbolic graph viewer
- Enables resource comparisons and integration
- Open development and utilization web-service
Description
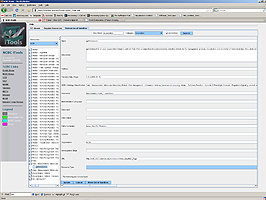
iTools is a meta-resource for managing the query, traversal and comparison of diverse computational biology resources. Specifically, iTools stores information about three types of resources–data, software tools and web-services. The iTools design, implementation and resource meta - data content reflect the broad research, computational, applied and scientific expertise available at the 7 National Centers for Biomedical Computing (www.NCBCs.org). iTools provides a system for classification, categorization and integration of different computational biology resources across space-and-time scales, biomedical problems, computational infrastructures and mathematical foundations. A large number of resources are already iTools-accessible to the community and this infrastructure is rapidly growing. iTools includes human and machine interfaces to its resource meta-data repository. Investigators or computer programs may utilize these interfaces to search, compare, expand, revise and mine meta-data descriptions of existent computational biology resources. We propose two ways to browse and display the iTools dynamic collection of resources. The first one is based on an ontology of computational biology resources, and the second one is derived from hyperbolic projections of manifolds or complex structures onto planar discs. iTools is an open source project both in terms of the source code development as well as its meta-data content. iTools employs a decentralized, portable, scalable and lightweight framework for long-term resource management. We demonstrate several applications of iTools as a framework for integrated bioinformatics. iTools and the complete details about its specifications, usage and interfaces are available at the iTools web pageiTools employs a new mechanism, Biositemaps (
Usage
iTools user Guide is available online at <a href="http://cms.loni.ucla.edu/iToolsUsage.aspx">http://cms.loni.ucla.edu/iToolsUsage.aspx.</a>
System Requirements
OS-Independent
- Version: v1.1 build 75
- Size: N/A
- OS: N/A
- Processor: Any
- Memory: 256 MB
- Software: Java 1.5+
Installation
iTools only requires a Java-enabled web-browser for its human interface.Purpose
iTools is used as a web-service via its flexible interfaces (http://cms.loni.ucla.edu/iToolsInterface.aspx):
* SOAP/XML Interface (Simple Object Access Protocol)
* WSDL (Web Services Description Language)
o Description of the iTools WSDL interface
* Applet/HTML, see versions section
* Java Web-Start