Big Data Neuroimaging Study Uncovers Shared Signature of Six Major Psychiatric Disorders
While neuroimaging is a valuable window into what happens inside the brain, it’s still an imprecise measure. Even after decades of technological advances, magnetic resonance imaging (MRI) scans still aren’t sharp enough to reveal what’s going on at the cellular level.
“By looking at brain scans, we don’t always know what mechanisms are driving a pattern of change in a given illness,” says Paul Thompson, PhD, professor of ophthalmology, neurology, psychiatry and the behavioral sciences, radiology, and engineering and associate director of the USC Mark and Mary Stevens Neuroimaging and Informatics Institute. “Researchers are left with a bit of a puzzle as to what those changes represent.”
But now, a revolutionary partnership between Thompson’s Enhancing Neuroimaging Genetics through Meta-Analysis (ENIGMA) consortium—a massive data-sharing effort between scientists in 45 countries—and researchers at the University of Toronto has led to a new method that can help pin down the mechanisms behind key psychiatric and developmental disorders. It’s a powerful breakthrough that bridges the gap between imaging and genetics research to identify the specific cells and molecules at play.
The team analyzed data on cortical thickness from more than 28,000 disease patients and healthy controls, ages 2 to 89, including individuals with schizophrenia, bipolar disorder, major depressive disorder, obsessive-compulsive disorder, autism spectrum disorder and attention-deficit/hyperactivity disorder. They identified a common molecular signature that spans all six disorders, publishing their findings in the journal JAMA Psychiatry in August.
The results can help pharmaceutical companies develop drug treatments that more directly address brain abnormalities, while the method developed by the team can be applied to other illnesses gain innumerable insights into the brain.
“This is a unique opportunity for science,” says Tomáš Paus, MD, PhD, professor at the University of Toronto and senior author of the paper. “ENIGMA has helped drive social innovation that allows us to move forward incredibly quickly—and to generate powerful new knowledge.”
LINKING IMAGING AND GENETICS
The new method developed by Paus and his team, known as “virtual histology,” uses atlases of gene expression to connect the dots between MRI scans and what’s happening in the brain at the cellular level. That’s possible because specific groups of genes mark certain types of cells in the brain.
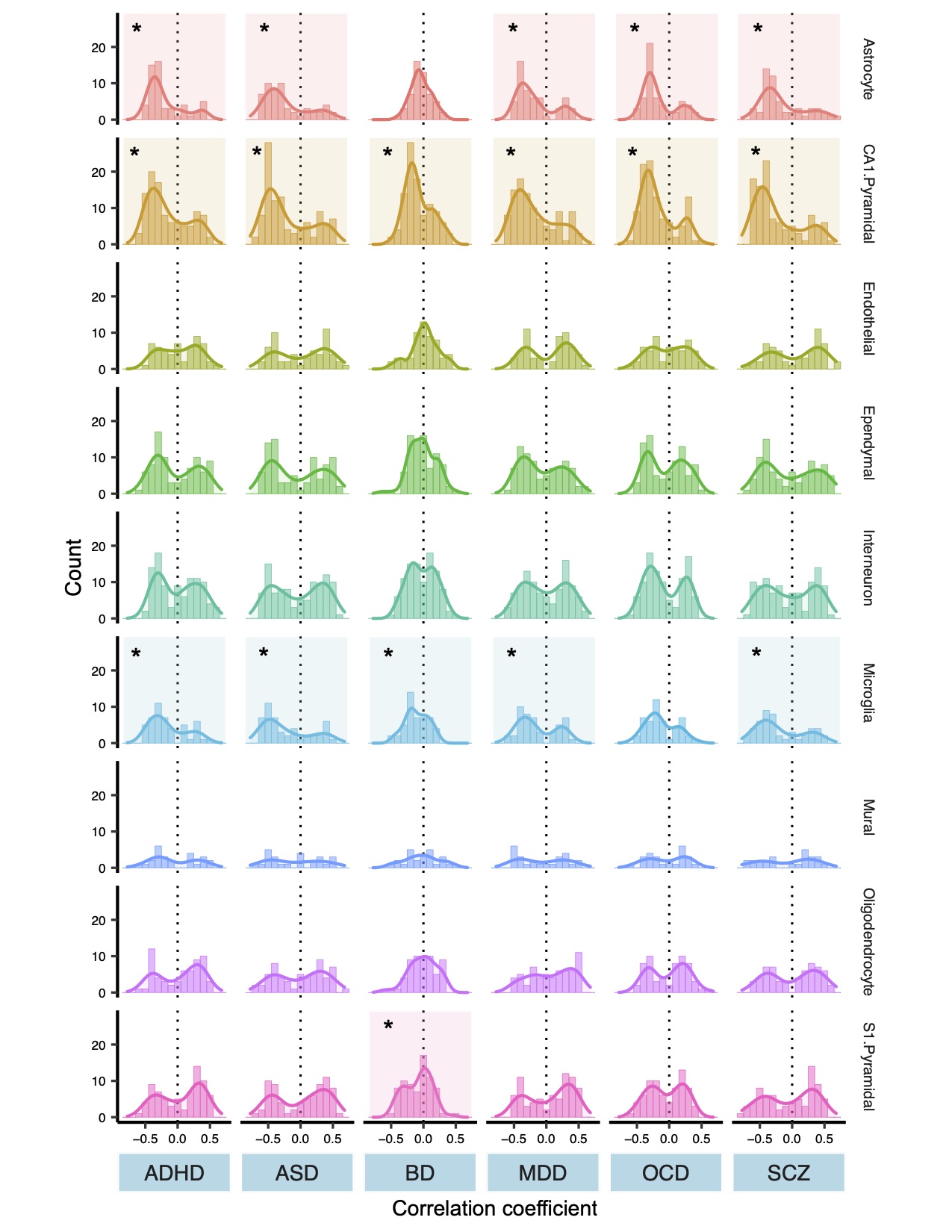
For example, imaging data revealed that one cell type in particular—the pyramidal neuron—often underlies the MRI signature of the diseases studied.
“Regions that have more of these cells show a bigger difference in cortical thickness between disease cases and controls,” Paus says.
Building on this finding, Yash Patel, first author of the paper and a doctoral student in Paus’ lab, conducted a “co-expression analysis,” which identifies the genes involved in shaping those cells and plots their expression over time.
“We found two groups of genes responsible for controlling those pyramidal cells—one prenatal and one postnatal,” says Patel.
The prenatal genes control the development of axons and other infrastructure in the brain, while the postnatal genes are related to plasticity and neurotransmission. Those findings suggest that people who develop schizophrenia, bipolar disorder and the other brain diseases studied may experience common environmental influences after birth that triggers disease onset, Patel says.
By applying the virtual histology method to the massive datasets aggregated by the ENIGMA network, the study’s authors have begun to uncover the biological mechanism that links genetic vulnerability for a disorder to actual changes in the brain.
NEXT STEPS
Looking forward, Paus and his team plan to conduct a similar analysis on the brain’s surface area and to apply the virtual histology method to a series of other problems, including Alzheimer’s disease, Parkinson’s disease and brain trauma.
“We can only develop practical remedies for these conditions if we know which genes, molecules and pathways to home in on,” Thompson says. “This method provides that crucial bridge for information to travel between the genetics and imaging communities.”